ΔG and PPB(LogIt) Prediction Model
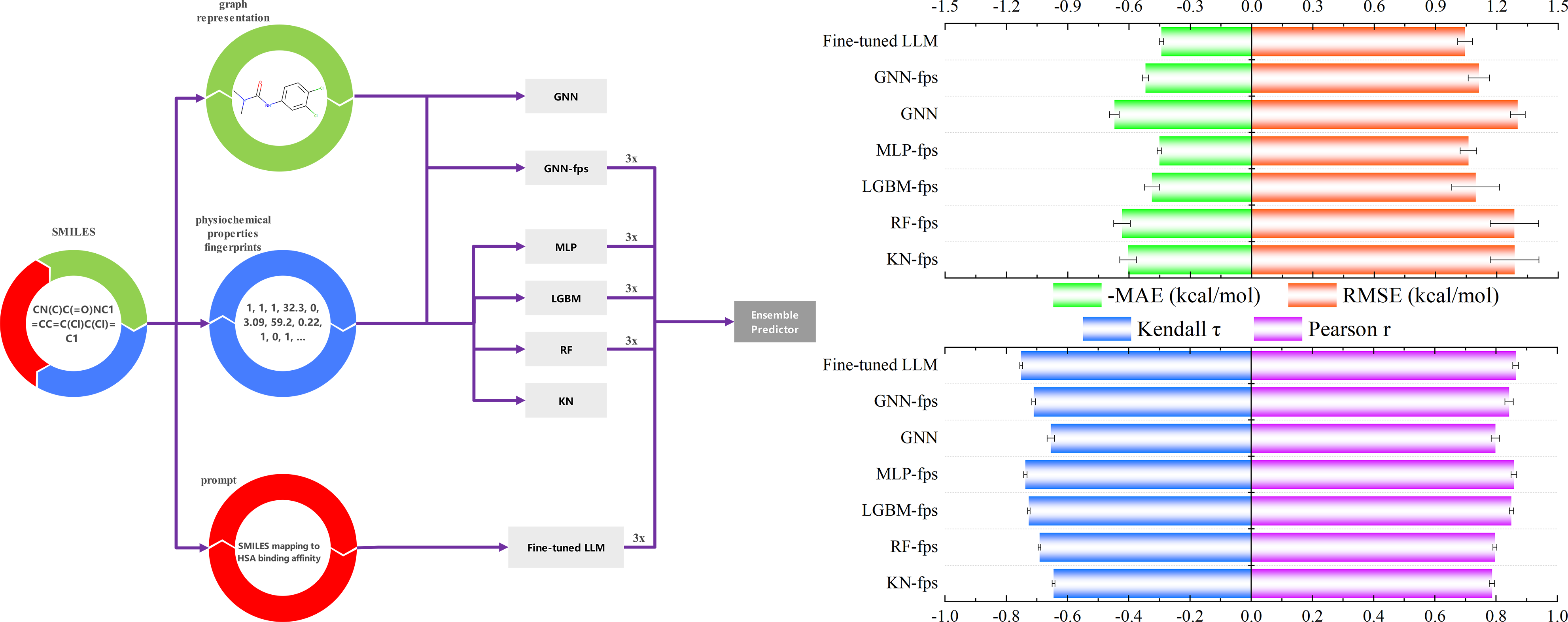
The affinity prediction module enables instant predictions of HSA-ligand binding affinities using ML predictors including MLP, LGBM, RandomForest and LLMs.
About PPB Prediction
Plasma Protein Binding (PPB) is a critical factor governing a drug's distribution and therapeutic potency. This module uniquely integrates pharmacophore-based fragmentation with advanced Graph Neural Networks (GNN) to capture key structural motifs. It provides precise predictions for HSA binding affinity (ΔG) and PPB, offering vital insights for ADME optimization.
Input Parameters
Structure Editor
Predicted Result
No results yet. Submit a SMILES structure and temperature.
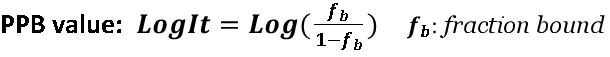
Physicochemical Properties
No properties calculated yet.
Distribution of Fragments
Fragment visualization will appear here after
prediction.